RxODE and rxode2
By Matthew Fidler in rxode2
October 12, 2022
RxODE vs rxode2
Since rxode2 came out recently, I am getting many questions about what is the difference between rxode2 and RxODE.
I think the biggest reason for the question is – is this update going to break all the nice things I already do with RxODE? Or maybe why should I bother to change?
I feel the same way when I have big changes in things I use. For me, I love the ability to pipe and change data with the tidyverse, and similar tools, but hate when they change things that affect my code.
With that in mind, I try to keep changes in behavior small when I modify things like RxODE and rxode2.
In this case, there were much more changes than usual and for that reason I wanted to change the name of the package to rxode2, but I believe most code will run well on either RxODE or rxode2. All changes to rxode2 are listed in the News/Changelog on rxode2’s website and is kept up to date.
What are the changes people may notice?
There are some changes that people will notice and may affect some code. In my opinion these are the big changes:
The options for
rxControlandrxSolveare more strict.camelCaseis now always used. Old options likeadd.covandtransit_absare no longer supported, onlyaddCovis supported. To me this is an annoyance and really makes things a bit easier to remember.The mnemonic
et(rate=model)andet(dur=model)mnemonics have been removed.rateneeds to be set to-1and-2manually instead.- This was done because the code for this was a bit cumbersome and hard to maintain
If you use options the prefix changed to
rxode2instead ofRxODE.- This was done so that
rxode2options will not breakRxODEoptions if you wish them to be different.
- This was done so that
Running simulations inside of an
rxode2block no longer depends on the number of threads used (a good fix that may be visible to some).By default the covariates are now added to the dataset (
addCov=TRUE) which is different than the behavior ofRxODE(addCov=FALSE).If you use transit compartments by
transit_absthis is no longer supported. Instead a specialevid=7is used by all transit compartment doses. This allows mixing with other types of doses into different compartments and better flexibility but will break code.
Things you are unlikely to notice or miss
- Various language options (like optionally requiring semi-colons at the end of statements, not allowing
<-for instance).
Why bother changing?
Simulating nlmixr2/rxode2 models directly
To me the biggest advantage to using rxode2 is you can simulate from nlmixr2 style models directly. For example, if you wanted to simulate the example nlmixr2 model you can use the following
library(rxode2)
set.seed(42)
rxSetSeed(42)
one.compartment <- function() {
ini({
tka <- 0.45 # Log Ka
tcl <- 1 # Log Cl
tv <- 3.45 # Log V
eta.ka ~ 0.6
eta.cl ~ 0.3
eta.v ~ 0.1
add.sd <- 0.7
})
# and a model block with the error sppecification and model specification
model({
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
d/dt(depot) = -ka * depot
d/dt(center) = ka * depot - cl / v * center
cp = center / v
cp ~ add(add.sd)
})
}
# Create an event table
et <- et(amt=300) %>%
et(0,24, by=2) %>%
et(id=1:12)
# simulate directly from the model
s <- rxSolve(one.compartment, et)## ℹ parameter labels from comments will be replaced by 'label()'print(s)## ── Solved rxode2 object ──
## ── Parameters ($params): ──
## # A tibble: 12 × 8
## id tka tcl tv add.sd eta.ka eta.cl eta.v
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 0.45 1 3.45 0.7 -0.206 -1.24 -0.690
## 2 2 0.45 1 3.45 0.7 -0.392 -0.0876 0.395
## 3 3 0.45 1 3.45 0.7 0.721 0.383 0.0316
## 4 4 0.45 1 3.45 0.7 -0.502 -0.0715 -0.221
## 5 5 0.45 1 3.45 0.7 0.791 0.295 0.488
## 6 6 0.45 1 3.45 0.7 -1.39 0.445 -0.665
## 7 7 0.45 1 3.45 0.7 -0.846 0.440 0.351
## 8 8 0.45 1 3.45 0.7 -0.700 0.740 0.275
## 9 9 0.45 1 3.45 0.7 1.87 -0.158 0.0480
## 10 10 0.45 1 3.45 0.7 0.281 -0.572 0.333
## 11 11 0.45 1 3.45 0.7 -1.86 0.159 0.450
## 12 12 0.45 1 3.45 0.7 -0.661 0.199 0.0133
## ── Initial Conditions ($inits): ──
## depot center
## 0 0
## ── First part of data (object): ──
## # A tibble: 156 × 10
## id time ka cl v cp ipredSim sim depot center
## <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 0 1.28 0.788 15.8 0 0 1.55 300 0
## 2 1 2 1.28 0.788 15.8 16.3 16.3 16.4 23.4 258.
## 3 1 4 1.28 0.788 15.8 16.1 16.1 15.4 1.82 254.
## 4 1 6 1.28 0.788 15.8 14.6 14.6 14.4 0.142 231.
## 5 1 8 1.28 0.788 15.8 13.3 13.3 12.9 0.0111 210.
## 6 1 10 1.28 0.788 15.8 12.0 12.0 11.9 0.000863 190.
## # … with 150 more rowsplot(s, center)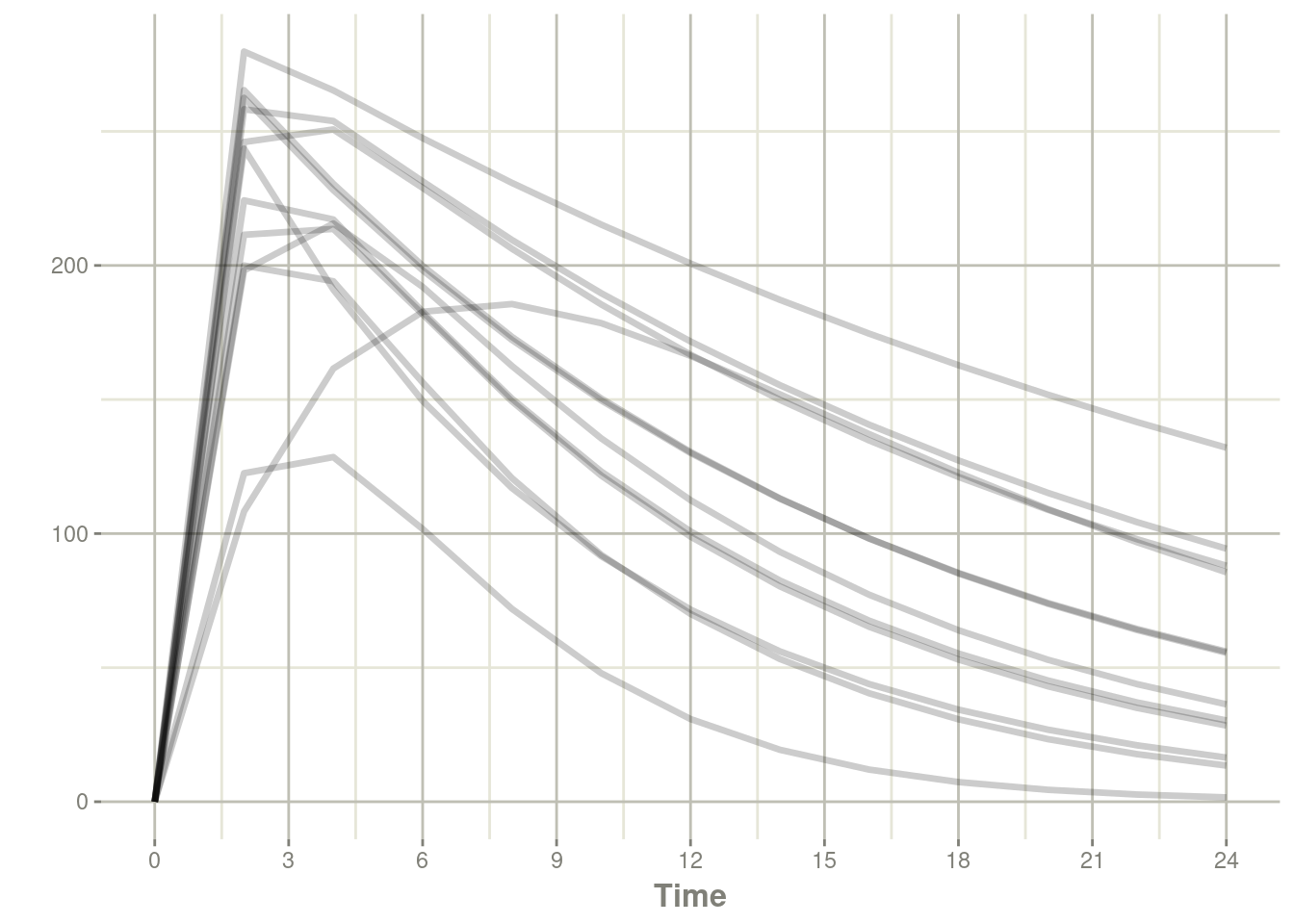
Notice that nlmixr2 was not called (or even required) to simulate this model.
Exploring and changing nlmixr2/rxode2 models directly
One of the nice features is you can change the model by simply changing a line or two of code by a feature called “model piping”. In the model piping included in rxode2 not only does it change your model it tells you how it is changed.
Lets assume you want to explore the impact of between subject variability in the model. You could drop the variability by changing a single line ka <- exp(tka+eta.ka) to ka <- exp(tka). Model piping allows that to occur easily
mod2 <- one.compartment %>%
model(ka <- exp(tka))## ℹ parameter labels from comments will be replaced by 'label()'## ! remove between subject variability `eta.ka`print(mod2)## ── rxode2-based free-form 2-cmt ODE model ──────────────────────────────────────
## ── Initalization: ──
## Fixed Effects ($theta):
## tka tcl tv add.sd
## 0.45 1.00 3.45 0.70
##
## Omega ($omega):
## eta.cl eta.v
## eta.cl 0.3 0.0
## eta.v 0.0 0.1
##
## States ($state or $stateDf):
## Compartment Number Compartment Name
## 1 1 depot
## 2 2 center
## ── μ-referencing ($muRefTable): ──
## theta eta level
## 1 tcl eta.cl id
## 2 tv eta.v id
##
## ── Model (Normalized Syntax): ──
## function() {
## ini({
## tka <- 0.45
## label("Log Ka")
## tcl <- 1
## label("Log Cl")
## tv <- 3.45
## label("Log V")
## add.sd <- c(0, 0.7)
## eta.cl ~ 0.3
## eta.v ~ 0.1
## })
## model({
## ka <- exp(tka)
## cl <- exp(tcl + eta.cl)
## v <- exp(tv + eta.v)
## d/dt(depot) = -ka * depot
## d/dt(center) = ka * depot - cl/v * center
## cp = center/v
## cp ~ add(add.sd)
## })
## }# simulate directly from the model
s <- rxSolve(mod2, et)
print(s)## ── Solved rxode2 object ──
## ── Parameters ($params): ──
## # A tibble: 12 × 7
## id tka tcl tv add.sd eta.cl eta.v
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 0.45 1 3.45 0.7 0.0771 0.782
## 2 2 0.45 1 3.45 0.7 -0.681 -0.170
## 3 3 0.45 1 3.45 0.7 0.457 -0.105
## 4 4 0.45 1 3.45 0.7 -0.336 -0.0738
## 5 5 0.45 1 3.45 0.7 -0.341 0.367
## 6 6 0.45 1 3.45 0.7 0.244 -0.411
## 7 7 0.45 1 3.45 0.7 0.212 -0.477
## 8 8 0.45 1 3.45 0.7 -0.332 0.135
## 9 9 0.45 1 3.45 0.7 -0.420 -0.439
## 10 10 0.45 1 3.45 0.7 0.0662 -0.313
## 11 11 0.45 1 3.45 0.7 -0.561 0.428
## 12 12 0.45 1 3.45 0.7 -0.146 -0.163
## ── Initial Conditions ($inits): ──
## depot center
## 0 0
## ── First part of data (object): ──
## # A tibble: 156 × 10
## id time ka cl v cp ipredSim sim depot center
## <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 0 1.57 2.94 68.8 0 0 1.28 300 0
## 2 1 2 1.57 2.94 68.8 3.92 3.92 3.53 13.0 270.
## 3 1 4 1.57 2.94 68.8 3.77 3.77 4.16 0.566 259.
## 4 1 6 1.57 2.94 68.8 3.47 3.47 3.58 0.0246 239.
## 5 1 8 1.57 2.94 68.8 3.18 3.18 3.58 0.00107 219.
## 6 1 10 1.57 2.94 68.8 2.92 2.92 2.80 0.0000463 201.
## # … with 150 more rowsplot(s, center)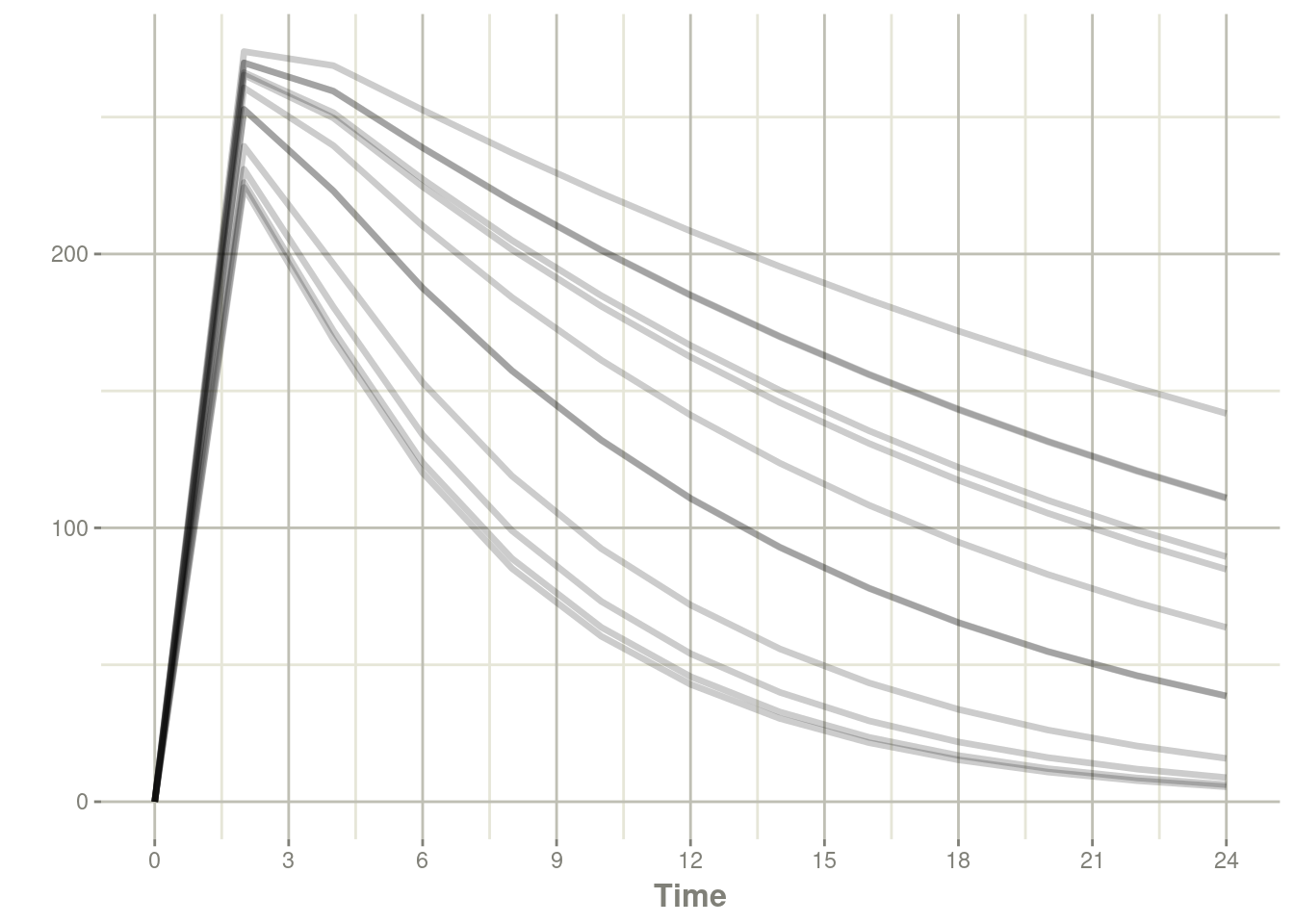
Not surprisingly without between subject variability on the ka component, there is not much difference in absorption between subjects.
What about piping a classic rxode2 model?
Well when rxode2 2.0.9 is released, you can also pipe a classic rxode2 model, which will change it to a nlmixr2 style model:
mod1 <- rxode2({
C2 <- centr/V2;
C3 <- peri/V3;
d/dt(depot) <- -KA*depot;
d/dt(centr) <- KA*depot - CL*C2 - Q*C2 + Q*C3;
d/dt(peri) <- Q*C2 - Q*C3;
d/dt(eff) <- Kin - Kout*(1-C2/(EC50+C2))*eff;
})
mod2 <- mod1 %>%
model(KA <- exp(tka+eta.ka),
append=NA) %>% # Prepend a line by append=NA
ini(tka = log(0.294),
eta.ka = 0.2,
CL = 18.6,
V2 = 40.2, # central
Q = 10.5,
V3 = 297, # peripheral
Kin = 1,
Kout = 1,
EC50 = 200) %>%
model(eff(0) <- 1)
print(mod2)
s <- rxSolve(mod2, et)
plot(s, centr, eff)
Why wouldn’t I want to switch?
rxode2 requires R 4.0. This is because the BH headers in windows require the R 4.0 toolchains to compile. To remain on CRAN, we needed to have the R 4.0 requirement.
While there may be ways to work-around this in Windows, the new version of rxode2 is not tested with R 4.0, and the old work-around was not straight forward. I cannot recommend you use rxode2 on any of the R versions before R 4.0; it would be too hard to reproduce.
So, in short, if you don’t have R 4.0, I wouldn’t recommend switching.