babelmixr2, nlmixr2 and Monolix
By Matt Fidler and the nlmixr2 Development Team in babelmixr2
December 5, 2022
As with NONMEM, it is important to be able to compare nlmixr2 to
industry standard software like Monolix. With that , in mind, I am
proud to announce the first nlmixr2 to Monolix translator in
babelmixr2.
As with NONMEM, while this has been done before, the method whereby we are converting between the two is novel and has some surprising advantages.
How to use Monolix with nlmixr2
To use Monolix in nlmixr, you do not need to change your data or your
nlmixr2 dataset. babelmixr2 will do the heavy lifting here.
Lets take the classic warfarin example to start the comparison with…
The model we use in the nlmixr2 vignettes is:
library(babelmixr2)
# The nonmem translation requires the package pmxTools as well.
# You do not need to load it, simply have it available for use.
pk.turnover.emax3 <- function() {
ini({
tktr <- log(1)
tka <- log(1)
tcl <- log(0.1)
tv <- log(10)
##
eta.ktr ~ 1
eta.ka ~ 1
eta.cl ~ 2
eta.v ~ 1
prop.err <- 0.1
pkadd.err <- 0.1
##
temax <- logit(0.8)
tec50 <- log(0.5)
tkout <- log(0.05)
te0 <- log(100)
##
eta.emax ~ .5
eta.ec50 ~ .5
eta.kout ~ .5
eta.e0 ~ .5
##
pdadd.err <- 10
})
model({
ktr <- exp(tktr + eta.ktr)
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
emax = expit(temax+eta.emax)
ec50 = exp(tec50 + eta.ec50)
kout = exp(tkout + eta.kout)
e0 = exp(te0 + eta.e0)
##
DCP = center/v
PD=1-emax*DCP/(ec50+DCP)
##
effect(0) = e0
kin = e0*kout
##
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl / v * center
d/dt(effect) = kin*PD -kout*effect
##
cp = center / v
cp ~ prop(prop.err) + add(pkadd.err)
effect ~ add(pdadd.err) | pca
})
}Next you have to either figure out the command to run Monolix, or
simply install the lixoftConnectors package
(https://monolix.lixoft.com/monolix-api/lixoftconnectors_installation/).
If the lixoftConnectors package is available, it will run Monolix
from R using that package.
If the way you run Monolix is a command, you can also set that command for the session by options("babelmixr2.monolix"="monolixRunCommand") or you can use it directly based on
monolixControl(runCommand="nmfe743"). This could also be a
function if you prefer (but I will not cover using the function here).
Lets assume you have lixoftConnectors or something similar setup appropriately. To run the
nlmixr2 model using Monolix you simply can run it directly:
fit <- nlmixr(pk.turnover.emax3, nlmixr2data::warfarin, "monolix",
monolixControl(modelName="pk.turnover.emax3"))## ℹ assuming monolix is running because 'pk.turnover.emax3-monolix.txt' is present## → loading into symengine environment...## → pruning branches (`if`/`else`) of full model...## ✔ done## → finding duplicate expressions in EBE model...## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → optimizing duplicate expressions in EBE model...## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → compiling EBE model...## ✔ done## → Calculating residuals/tables## ✔ done## → compress origData in nlmixr2 object, save 27560## ℹ monolix parameter history needs expoted charts, please export chartsThis fit issues an informational tidbit -
- monolix parameter history needs expoted charts, please export charts
This will automatically be generated as well when lixoftConnectors
package is generated and you have a recent version of Monolix. If you
don’t have that information then the important parameter history plots will
not be imported and you cannot see those plots.
Just like with the NONMEM translation, the monolixControl() has
modelName which helps control the output directory of Monolix (if
not specified babelmixr2 tries to guess based on the model name
based on the input).
Printing this out this nlmixr2 fit you see:
fitOf particular interest is the comparison between Monolix predictions and nlmixr predictions:
IPRED relative difference compared to Monolix IPRED: 0.09%; 95% percentile: (0.01%,0.49%); rtol=0.000941
PRED relative difference compared to Monolix PRED: 0.04%; 95% percentile: (0%,0.2%); rtol=0.000428
IPRED absolute difference compared to Monolix IPRED: atol=0.00911; 95% percentile: (0.000493, 0.0928)
PRED absolute difference compared to Monolix PRED: atol=0.000428; 95% percentile: (3.14e-07, 0.203)In this case, I believe that these also imply the models are
predicting the same thing. Note that the model predictions are not as
close as they were with NONMEM because Monolix does not use the
lsoda ODE solver. Hence this small deviation is expected, but still
gives a validated Monolix model.
As in the case of NONMEM, this gives some things that are not
available to Monolix, like adding conditional weighted residuals:
fit <- addCwres(fit)## → Calculating residuals/tables## ✔ doneWhich will add nlmixr’s CWRES as well as adding the nlmixr2 FOCEi objective function
OBJF AIC BIC Log-likelihood Condition Number
FOCEi 1335.312 2261.007 2340.427 -1111.503 2203.836
monolix 1522.704 2448.398 2527.819 -1205.199 2203.836Because you now have an objective function compared based on the same assumptions, you could compare the performance of Monolix and NONMEM based on objective function.
To be fair, objective function values must always be used with caution. How the model performs and predicts the data is far more valuable.
Also since it is a nlmixr2 object it would be easy to perform a VPC
too (the same is true for NONMEM models):
v1s <- vpcPlot(fit, show=list(obs_dv=TRUE), scales="free_y") +
ylab("Warfarin Cp [mg/L] or PCA") +
xlab("Time [h]")
v2s <- vpcPlot(fit, show=list(obs_dv=TRUE), pred_corr = TRUE, scales="free_y") +
ylab("Prediction Corrected Warfarin Cp [mg/L] or PCA") +
xlab("Time [h]")
v1s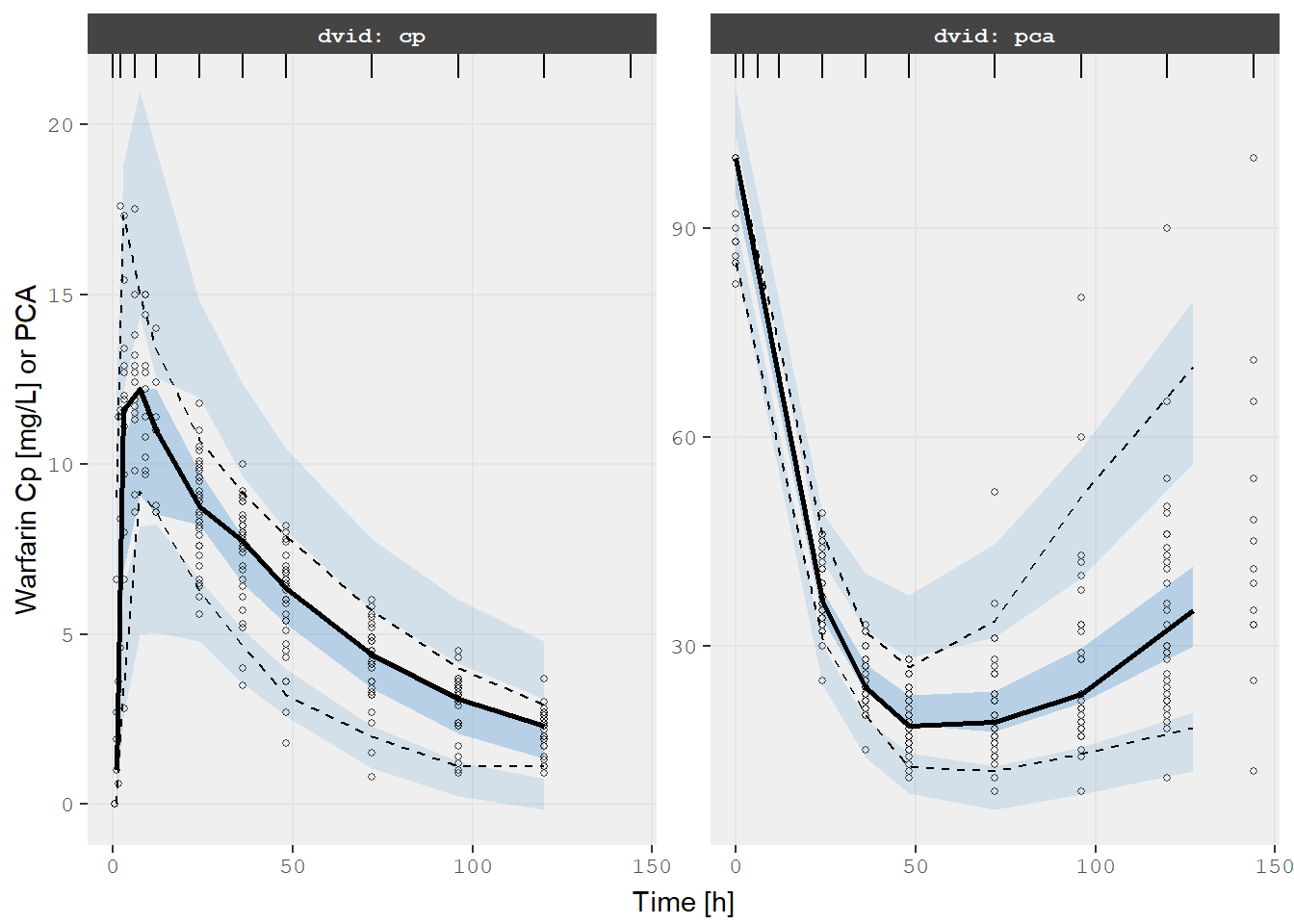
v2s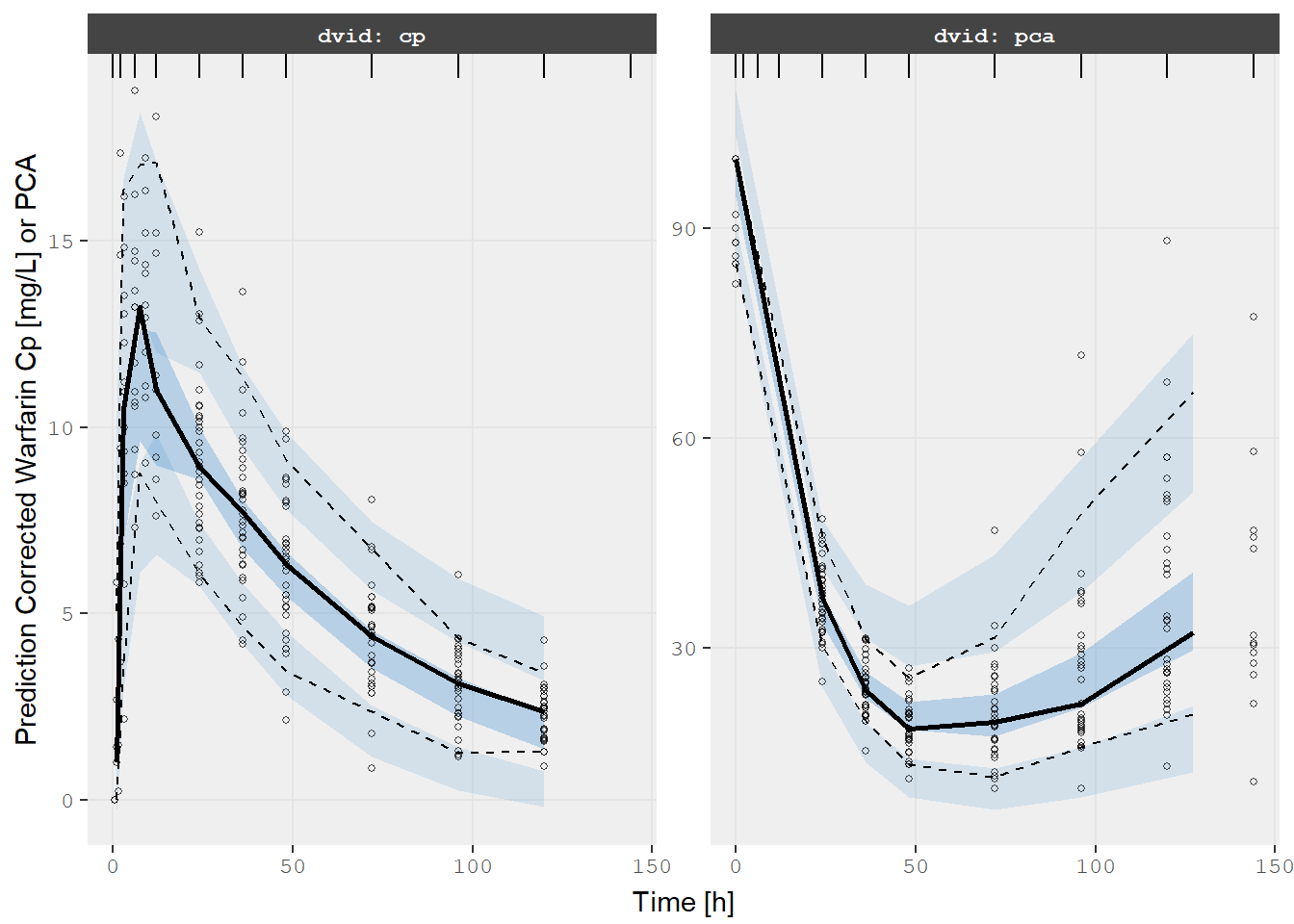
Note about data
The input dataset expected to be compatible with rxode2 or
nlmixr2. This dataset is then converted to Monolix format:
The combination of
CMTand Dose type creates a uniqueADMvariable.The
ADMdefinition is saved in the monolix model filebabelmixr2creates a macro describing the compartment, iecompartment(cmt=#, amount=stateName)babelmixr2also creates a macro for each type of dosing:Bolus/infusion uses
depot()and adds modeled lag time (Tlag) or bioavailibilty (p) if specifiedModeled rate uses
depot()withTk0=amtDose/rate.babelmixr2also adds modeled lag time (Tlag) or bioavailibilty (p) if specifiedModeled duration uses
depot()withTk0=dur, also add adds modeled lag time (Tlag) or bioavailibilty (p) if specified Turning off a compartment uses empty macro
- Posted on:
- December 5, 2022
- Length:
- 5 minute read, 1000 words
- Categories:
- babelmixr2
- Tags:
- new-version Monolix